Being stuck in an MRI scanner can be very boring. Consequently in many settings participants’ vigilance may decline during the scan. This is typically not a problem for structural scans. However, when we study functional correlation between brain regions at rest (dubbed functional connectivity) it does matter a great deal whether the participant is awake or asleep. The transition from wakefulness to sleep is characterized by profound changes in brain activity and also in functional correlation.
Previously we have published the development of a support-vector machine based classifier that can predict sleep stages from rs-fMRI data (paper at NeuroImage or a preliminary poster for OHBM 2013). This model uses parallel recorded EEG data as a gold-standard for sleep scoring. The MATLAB code for the model is available here at github.
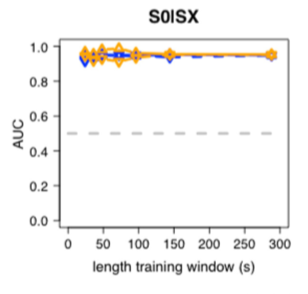
Classifier performance in AUC between wakefulness (S0) and sleep (SX) for training data (cross-validation; blue) and independent test data (orange). 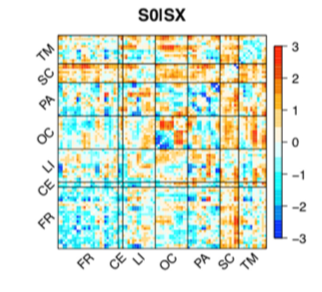
Feature weights in the linear SVM for the wakefulness (S0) vs sleep (SX) classifier.
Despite having tested the classifier on an independent test set (acquired at the same center) and also in people before and after administering propofol at a different center, the question was still how does the classifier perform in general. And can the results be biased (shifted) by technical scanner difference such as field strength or TR and also by demographics (e.g., age or sex) or instructions (eyes open vs eyes closed). In order to address this question we processed the resting state fMRI data of the 1000 functional connectomes dataset and systematically evaluate these potential sources of bias. The results are presented at OHBM 2019 (poster).