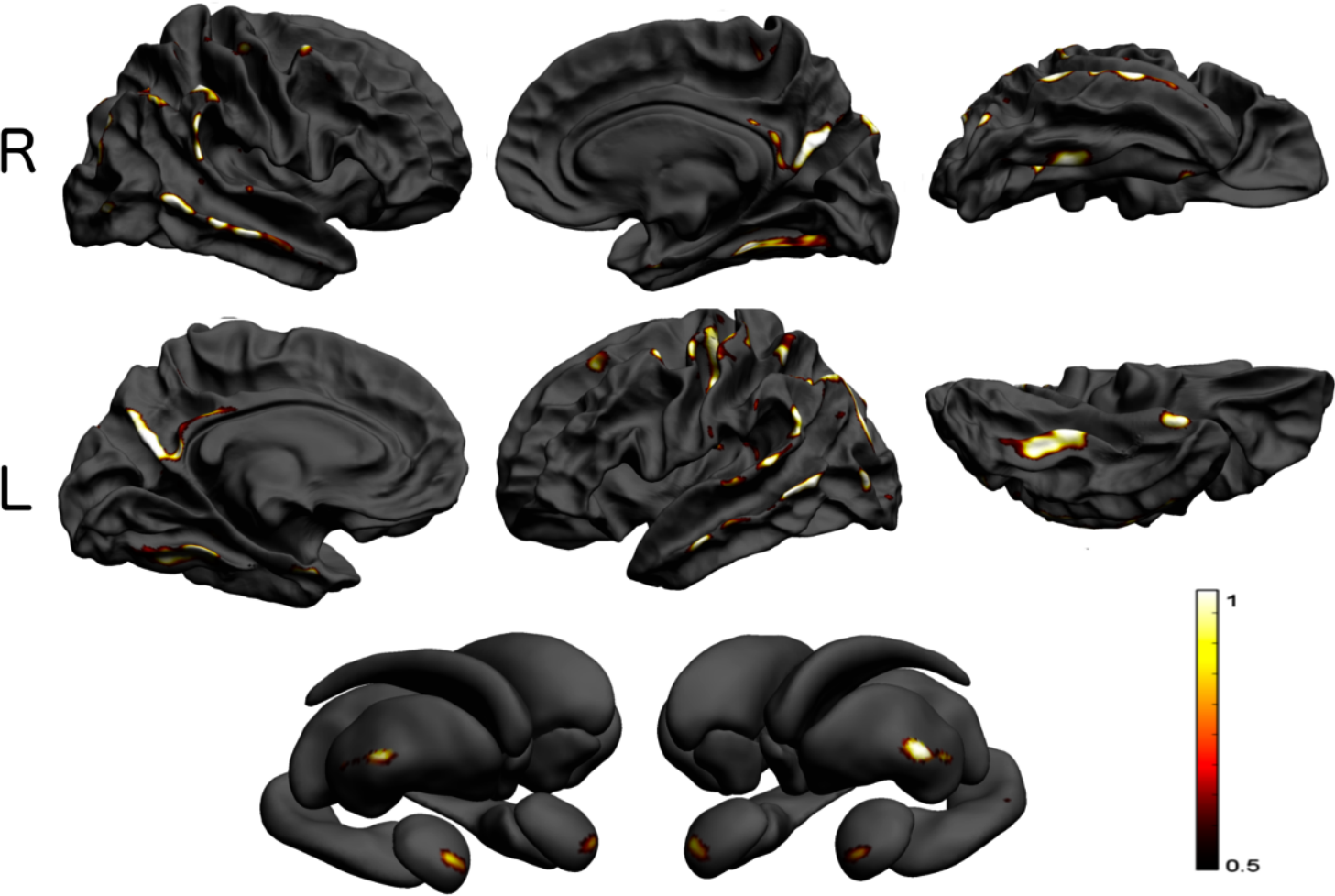
Do we need adjustment for population structure in neuroimaging?
Any large genome-wide association study corrects for population structure, in addition to restricting the sample to subjects of caucasian ethnicity. This is for the most part a practical necessity since mankind's…